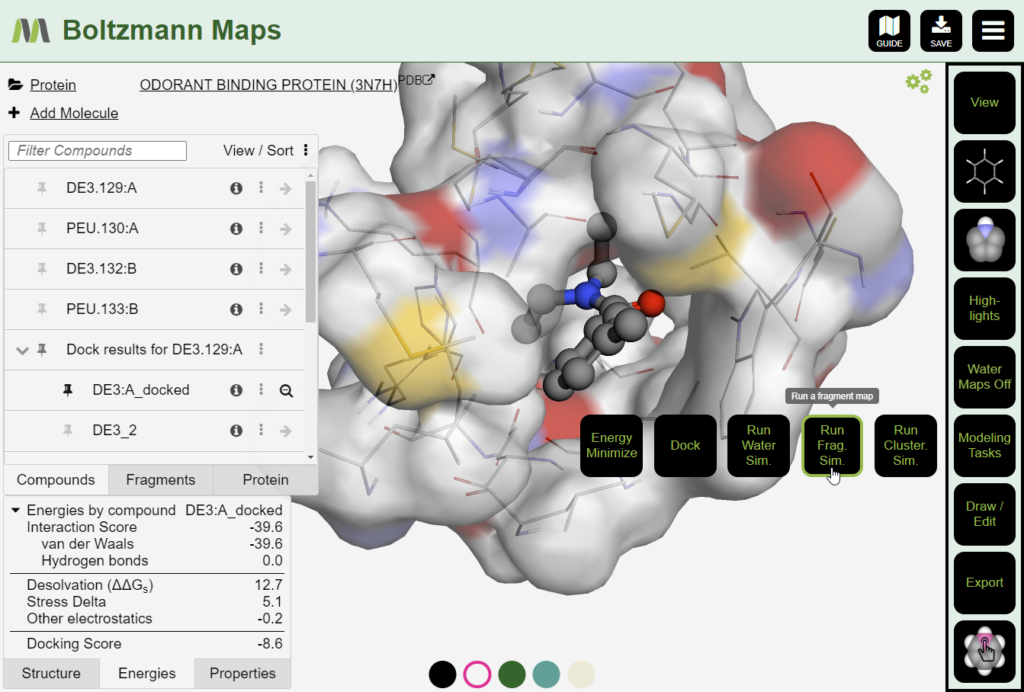
With the new release of Boltzmann Maps comes enhanced reliability for protein structure loading and automation of protein preparation for energy minimization, docking and fragment simulations. The entirety of the Protein Data Bank (PDB) is now available to view in Boltzmann Maps.
As an example, log into BMaps to view PDB ID 3n7h: https://www.boltzmannmaps.com/structure/3n7h. The PDB featured this mosquito odorant binding protein in complex with DEET (DE3 ligand) as one of the “Molecules of the Month” for June 2023.
Along with the ability to import directly from PDB, all small molecules are automatically assigned forcefield parameters and partial charges. We tested almost 200,000 entries from the PDB for successful accessibility in this release. Although all structures can load, obtaining forcefield parameters fails on a small number of cases due to errors in the PDB source. We are working with the PDB to have these corrected. Computing partial charges uses the AM1 semi-empirical method with the AM1-BCC corrections (via AmberTools). In cases where AM1 fails, PM6 is now used as a backup to handle more cases.
Unlike structures imported from the PDB, Boltzmann Maps prepared structures are always ready for computation. Our science team continues to optimize and validate new structures with enhanced protein preparation.
Behind the scenes protein preparation improvements
The improved coverage of PDB ligands is facilitated by the new mmCIF version of the PDB Chemical Component Dictionary, which contains bond order information for ligands in PDB structure files. The new format improves on the older het_dictionary.txt, resulting in more reliable preparation of forcefield parameters for these ligands.