Fragment maps for ten SARS-CoV-2 virus proteins are now available in the BMaps web app to accelerate the design of COVID-19 therapeutics. These structures include the main protein protease (NSP5), the Spike protein (S), the receptor binding domain (RDB) of the S protein, and several NS (non-structural) proteins NSP3, NSP9, NSP10, NSP15, NSP16. Available for each protein are druggability sites, water molecule maps, and a starting set of 117 chemical fragment binding maps.
Using an automated search tool, the maps are queried to identify fragments, ranked by affinity. These fragments bind to the protein in a conformation suitable for bond formation with a bound starting compound. Starting points may be co-crystal ligands, library compounds docked at druggable sites, or scaffolds selected from fragment maps. A new compound is constructed by growing or substituting a selected fragment at functional sites on the starting compound. Finally, the interaction energy of the new compound is reported to evaluate the potential for improved binding affinity.
This process can provide new ideas for rapidly moving your antiviral inhibitors to the clinic. To get started, view a SARS-CoV-2 structure now. Or login for free access to simulation features like docking and fragment growing. The Conifer Point team is willing to help. Contact us at john.kulp@coniferpoint.com.
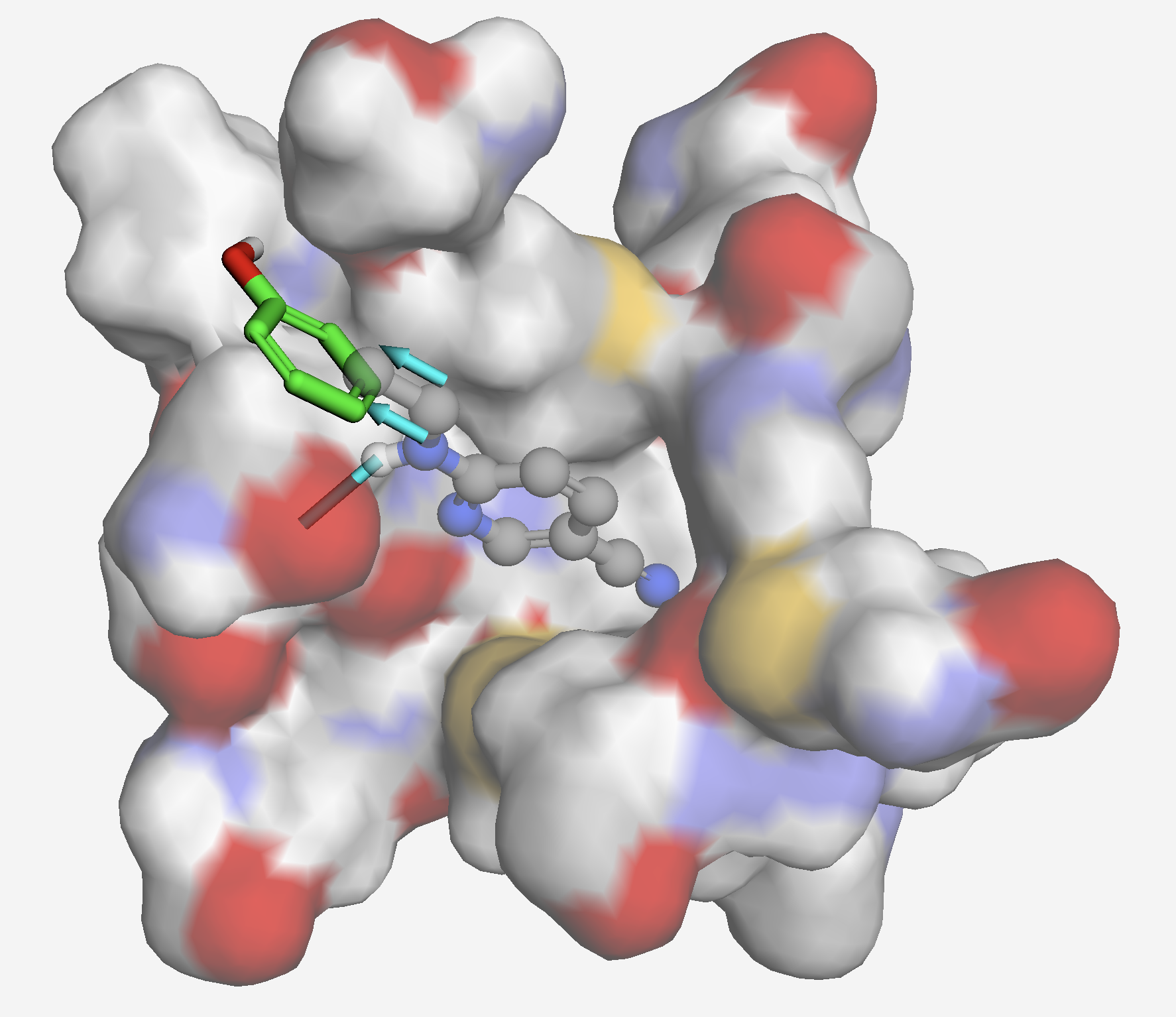
Crystal Structure of SARS-CoV-2 main protease in complex with Z219104216 (PDB: 5R82). Fragment growing reveals an opportunity with phenol to access an additional sub-pocket to increase affinity.
