Structure-based drug design starts with a compound positioned on the surface of a protein. Often, the crystal ligand in a PDB structure provides the natural starting point. But what if there is no ligand? This question took on increased urgency as the Boltzmann Maps team prepared fragment simulations on COVID-19 structures; many of the initial entries in the PDB did not have ligands. In response to this, Boltzmann Maps now provides Sample Compounds. We docked 20K+ small molecules from our libraries against hotspots on each structure and selected a handful to show in BMaps. The resulting compounds are generally commercially available, chemically diverse, and are reasonable starting points for structure-based design. They can then be used to explore new designs, using BMaps’ energy analysis and fragment data.
Sample Compounds are available for almost all of the SARS-CoV-2 structures in Boltzmann Maps. More are coming!
View a COVID-19 structure with sample compounds now >>. Or, log in to start exploring your own structure-based design modifications.
Using Sample Compounds for structure-based design
Loading sample compounds is easy; if they are available for your structure, you will see an “Add sample compounds” button. If you have your own compounds to work from, you can dismiss the option and use “Add Molecule” to add them.
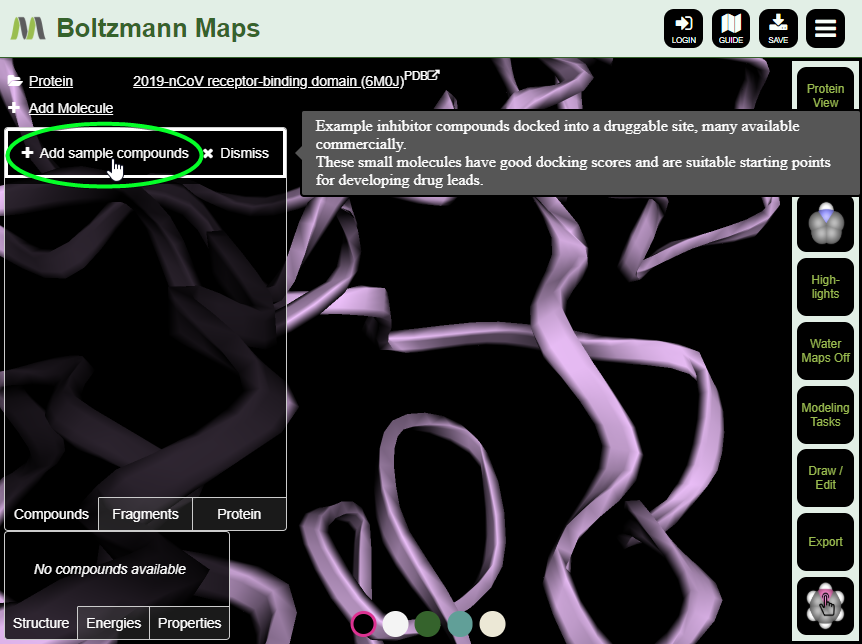
Once you have compounds in place, you can start making modifications. Switch to Ligand View, then use the right-click menu on compound atoms to perform terminal replacement or fragment growing. You can also use the 2D editor to make changes to compounds or draw new ones. After making changes, view the energy and properties tables to see whether the binding properties have improved. We’ll cover these kinds of modifications in more detail in a followup blog post.
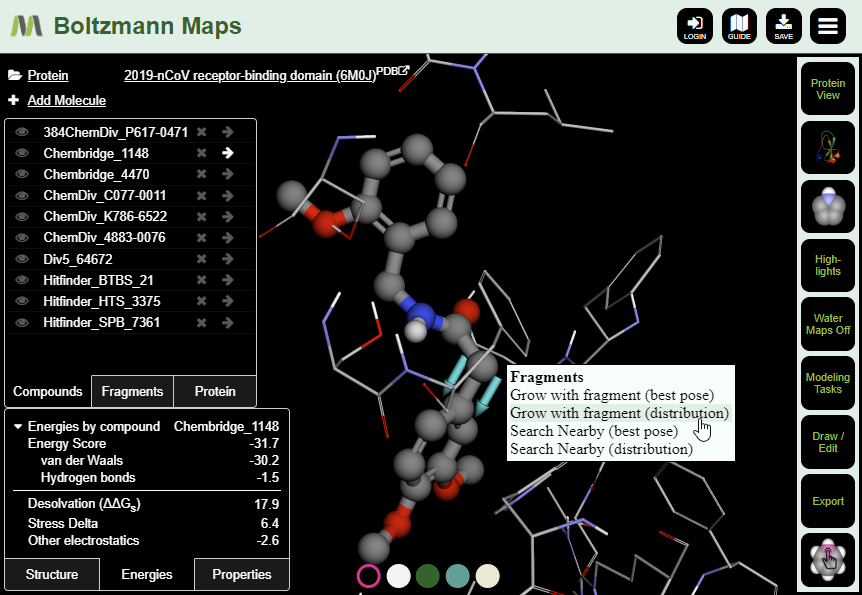
To get started with your own structure-based design modifications for COVID-19 targets, log in to Boltzmann Maps.
