Boltzmann Maps is pleased to introduce an integration with Pharmit as an option for pharmacophore screening. Pharmit is a search tool for finding small molecule inhibitors that bind to a target of interest. The tool searches libraries for compounds with desired features in the right geometry. Boltzmann Maps integration allows the user to send a protein-ligand system from BMaps to Pharmit for search based on the compound’s features or other user-specified features. Pharmit’s nine built-in libraries include almost 250M compound entries, and the 1,059 publicly accessible user-contributed libraries contain another 45M entries.
Pharmit can be accessed via the Export button on the bottom right of the BMaps web app.
Continue reading Pharmacophore screening using Pharmit in Boltzmann Maps →
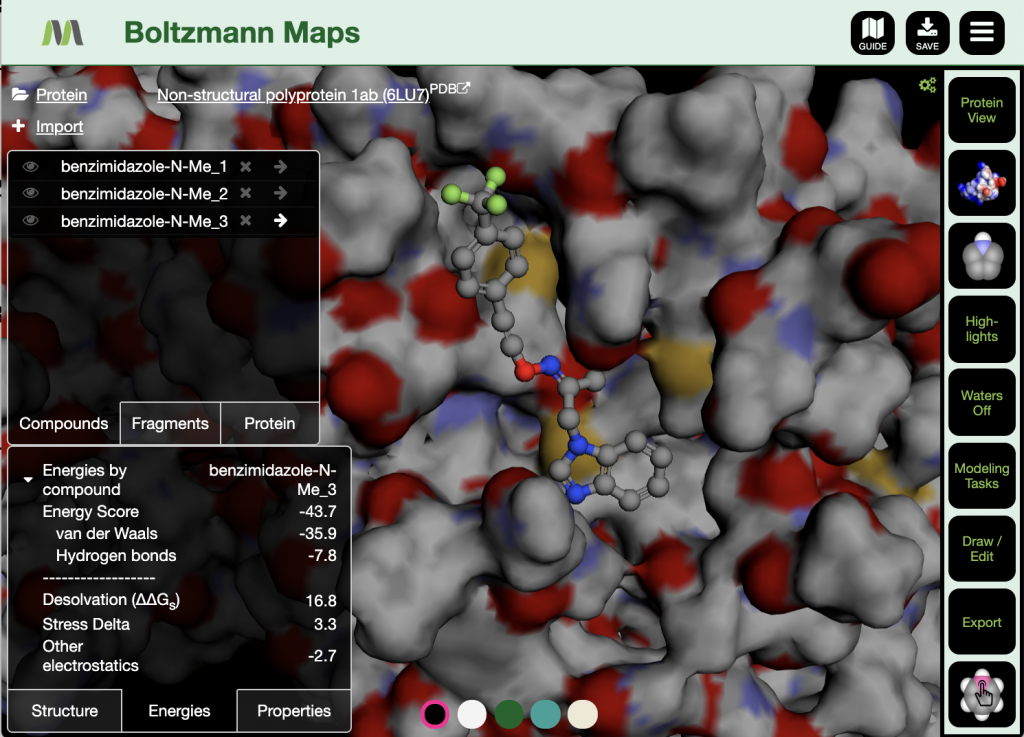

 BMaps version 1.0 is now released at the boltzmannmaps.com website, launching the first Web-based fragment-based design application using Boltzmann Maps.
BMaps version 1.0 is now released at the boltzmannmaps.com website, launching the first Web-based fragment-based design application using Boltzmann Maps.