Technology Behind BMaps
The BMaps web application incorporates technologies from an earlier company, BioLeap, Inc., and integrates numerous state-of-the-art molecular modeling software libraries and Web technologies:
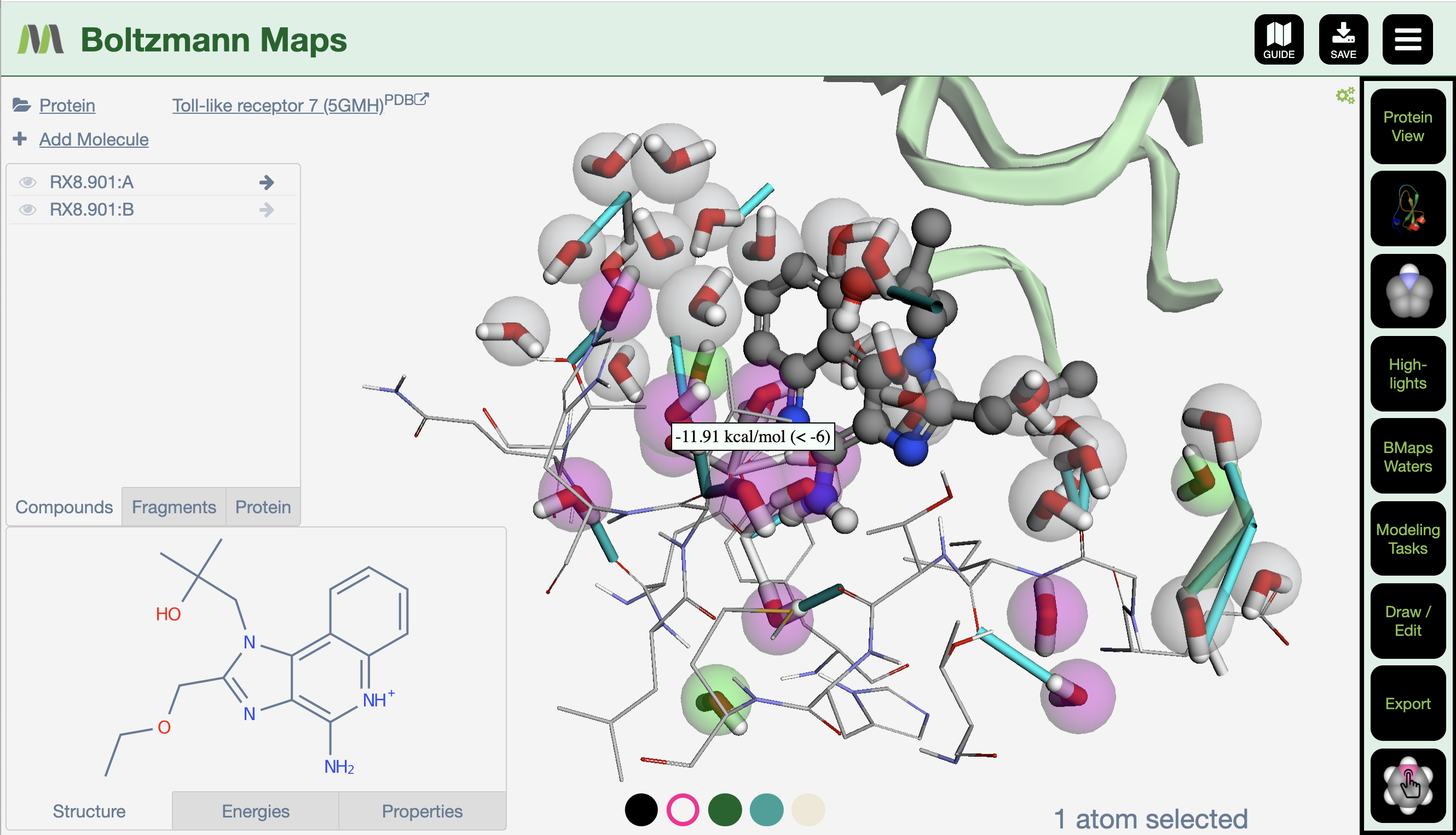
- BMaps web app
- 3Dmol.js for molecular visualization using WebGL (https://3dmol.csb.pitt.edu/)
- Ketcher 2D chemistry editor (https://lifescience.opensource.epam.com/ketcher/index.html)
- SmilesDrawer 2D chemistry structure displayer (https://github.com/reymond-group/smilesDrawer)
- BMaps AWS-hosted server program
- Derived from BioLeap’s BFD fragment-based design tools, including assignment of Amber forcefield parameters, energy minimization, chemical fragment searches, compound modification, and services integration
- LS-Align molecular alignment (https://zhanglab.ccmb.med.umich.edu/LS-align )
- Openbabel for molecular file format conversion (https://github.com/openbabel/openbabel)
- RDKit for 2D to 3D molecule conformation generation (https://www.rdkit.org/)
- OpenMM a high performance toolkit for molecular simulation, used for energy minimization (https://openmm.org)
- Tcl scripting language interpreter built-in, supporting a large body code from the BFD application (http://tcl.tk/)
- Fragment Simulations
- BMOC (Boltzmann Monte Carlo) Grand Canonical Monte Carlo with simulated annealing of chemical potential simulator
- BMaps Web Services
- The Indigo Toolkit enables searching fragment libraries—including CHEMBL—by drawn structures
- BMaps’s advanced tool chain supports user-launched computation of water and fragment maps on the user’s own structures.
- Other software
- AmberTools for deriving partial charges (https://ambermd.org/AmberTools.php)
- AutoDock Vina for molecular docking (http://vina.scripps.edu/)
- CPAWS (ConiferPoint AWS job management system)
Research projects are being pursued using WebSpeech for voice-controlled molecular visualization (see the microphone icon on the development web site https://lab.boltzmannmaps.com/).